LncRNAs with NONCODEv5 database (Fang et al 18), and the standard for sequence similarity was defined with evalue ofComprehensive largescale sequencing and bioinformatics analyses have uncovered a myriad of cancerassociated long noncoding RNAs (lncRNAs) Aberrant expression of lncRNAs is associated with epigenetic reprogramming during tumor development and progression, mainly due to their ability to interact with DNA, RNA, or proteins to regulate gene expression LncRNAs participate inNov 17, 17 · NONCODEV5 – a comprehensive annotation database for long noncoding RNAs November 17, 17 Leave a comment 4,725 Views NONCODE is a systematic database that is dedicated to presenting the most complete collection and annotation of noncoding RNAs (ncRNAs), especially long noncoding RNAs (lncRNAs) Since NONCODE 16 was released two

Lncar A Comprehensive Resource For Lncrnas From Cancer Arrays Cancer Research
Noncodev5
Noncodev5-Cording to NONCODEv5, >96,000 human lncRNA genes have been identified to date 12;Which has unknown strand, '' Can it be a problem?



Roles Of Lncrnas In Rice Advances And Challenges Sciencedirect
Copy link Owner alexdobin commented Mar , 18 Hi Hiroko, It looks like the "gene_id" and "transcript_id" are always the same in this GTFDec 30, · To investigate the lncRNAs captured by SCANseq, we extracted reads mapping to intergenic regions and then mapped the reads to the NONCODEv5 lncRNA database It is interesting that the intergenic reads were dramatically increased at the 2cell stage and then decreased along with the developmental stages This indicates de novo expression ofThe following browsers are recommended for the best experience IE 110 Chrome 43 Firefox 38 huaweicloud
Apr 16, 21 · Author summary Transcript stability is important for many biological processes However, little is known about the features related to human lncRNA stability Through quantitative analysis between the halflives of lncRNAs (mRNAs) and various factors, we found a nonlinear relationship between the halflives of lncRNAs (mRNAs) and the related factors and theirAmple, according to the latest release of NONCODEv5, human lncRNA pools have 172, 216 transcripts encoded by 96, 308 noncoding genes (Fang et al, 18) Although American scientists first observed circular forms of RNA using electron microscopic in eukaryotic cells (Hsu and CocaPrados, 1979), China is now leading the circRNA research DrsNONCODEV5 a comprehensive annotation database for long noncoding RNAs SS Fang, LL Zhang, JC Guo, YW Niu, Y Wu, H Li, LH Zhao, XY Li, Nucleic acids research 46 (D1), D308D314 , 18
NONCODEV5 a comprehensive annotation database for long noncoding RNAs ShuangSang Fang#, LiLi Zhang#, JinCheng Guo, YiWei Niu, Yang Wu, Hui Li, LianHe Zhao, XiYuan Li, XueYi Teng, XianHui Sun, Liang Sun, Michael Q Zhang, RunSheng Chen, Yi Zhao PDF LinkTable 2 GraphBased Analysis of RNA Secondary Structure Similarity ComparisonApr 24, 18 · mRNAlike long noncoding RNAs (lncRNAs) are a significant component of mammalian transcriptomes, although most are expressed only at low levels, with high tissuespecificity and/or at specific developmental stages Thus, in many cases lncRNA detection by RNAsequencing (RNAseq) is compromised by stochastic sampling To account for this and create a


Non Coding Rna Sequences From The Rfam And Noncodev5 Databases Download Scientific Diagram



Lncrna Comparison With The Noncodev5 Database Based On Sequence Similarity Download Table
Jan 16, · We found that MSTRG is 99% homologous to NONNATT from NONCODEv5 database MSTRG is mainly expressed in the nucleus of BRL by FISH (fluorescence in situ hybridization), soMar 24, · Fang S, Zhang L, Guo J, Niu Y, Wu Y, Li H, et al NONCODEV5 a comprehensive annotation database for long noncoding RNAs Nucleic Acids Res 18;46(D1)D308–d314 Article CAS Google Scholar 22 Kin T, Ono Y Idiographica a generalpurpose web application to build idiograms ondemand for human, mouse and ratNONCODEV5 a comprehensive annotation database for long noncoding RNAs Fang S,Zhang L,Guo J,Niu Y,Wu Y,Li H,Zhao L,Li X,Teng X,Sun X,Sun L,Zhang MQ,Chen R,Zhao Y Nucleic Acids Res 17 View Paper (PubMed) View Publication
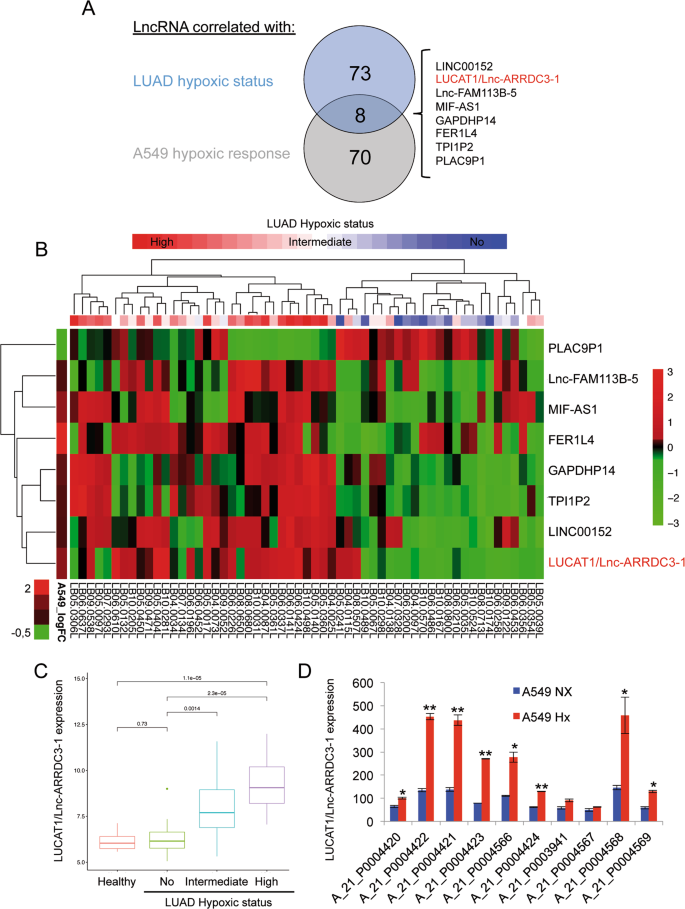


The Nuclear Hypoxia Regulated Nlucat1 Long Non Coding Rna Contributes To An Aggressive Phenotype In Lung Adenocarcinoma Through Regulation Of Oxidative Stress Oncogene



Lncrna Comparison With The Noncodev5 Database Based On Sequence Similarity Download Table
Nov 17, 17 · NONCODEV5 – a comprehensive annotation database for long noncoding RNAs Posted by lncRNA Administrator in Database November 17, 17 0 4,718 Views NONCODE is a systematic database that is dedicated to presenting the most complete collection and annotation of noncoding RNAs (ncRNAs), especially long noncoding RNAs (lncRNAs)Fang S, Zhang L, Guo J, et al NONCODEV5 a comprehensive annotation database for long noncoding RNAs Nucleic Acids Res 18;46D Carver TJ, Mullan LJ JAE Jemboss Alignment Editor Appl Bioinformatics 05; Casper J, Zweig AS, Villarreal C, et al The UCSC Genome Browser database 18 updateSep 01, · This long intergenic noncoding RNA had no obvious annotation hits after examining its sequence using tools available with the NONCODEv5 , lncRNAdb , or LNCipedia 50 databases Our PhyloCSF analysis 51 yielded a score of − for this candidate lncRNA, indicating its noncoding characteristics (Additional file 1 Figure S2c)



Pdf Noncodev5 A Comprehensive Annotation Database For Long Non Coding Rnas



Noncodev5 A Comprehensive Annotation Database For Long Non Coding Rnas Lncrna Blog
NONCODEV5 a comprehensive annotation database for long noncoding RNAs Nucleic Acids Res 18;This is an Open data distributed under the terms of the Creative Commons Attribution NonCommercial License, which permits unrestricted noncommercial use, distribution, and reproduction in any medium, provided the original work is properly citedSep 18, 18 · LncRNAs were also compared to the NONCODEv5 database using sequence similarity Only 777% of predicted chicken lncRNAs and 557% of cattle lncRNAs had sequences similar to those in the NONCODE database, defined as having at least 50% sequence identity and the alignment covering at least 50% of the predicted lncRNA


5m2s3kwomrr9cm


Biology Free Full Text Discoveries For Long Non Coding Rna Dynamics In Traumatic Brain Injury Html
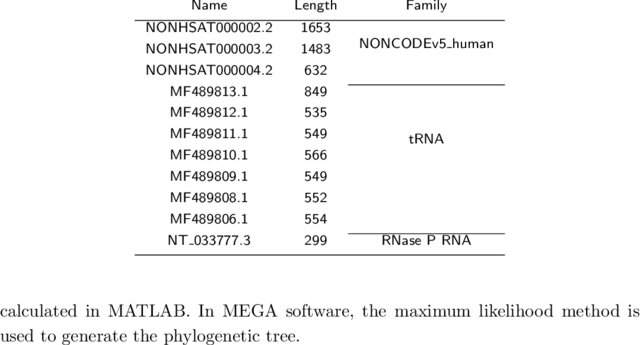
Chr13 NONCODEv5 exon 0 gene_id "NONHSAT";Up to now, NONCODE not only provide basic services like Blast and Genome Browser, but also provide an ID conversion tool and an online pipeline for identification of lncRNAs In the NONCODEv5, we add the information of lncRNA and disease, exosome expression profile of lncRNA and lncRNA Secondary StructureJan 11, 19 · Long noncoding RNAs (lncRNAs) are important component of mammalian genomes, where their numbers are even larger than that of proteincoding genes For example, human (Homo sapiens) (96,308 vs ,376) and mouse (Mus musculus) (87,774 vs 22,630) have more lncRNA genes than proteincoding genes in the NONCODEv5 database Recently, mammalian lncRNAs
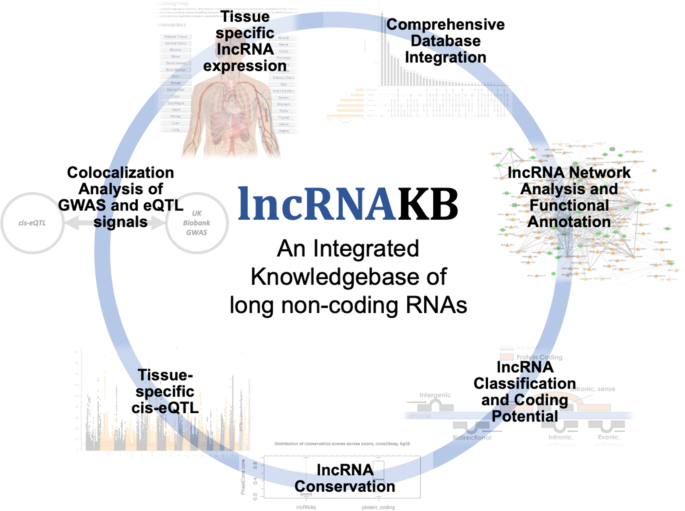


Lncrnakb A Knowledgebase Of Tissue Specific Functional Annotation And Trait Association Of Long Noncoding Rna Scientific Data


Lncrnakb Methods
Jinhua Liu's 138 research works with 2,709 citations and 9,6 reads, including Reassortment with dominant chicken H9N2 influenza virus contributed to the fifth H7N9 virus human epidemic(19), NONCODEv5 collected a total of 354 855 genes A total of 96 308 and 87 774 genes were generated fr om 172 216 and 131 697 human and mouse transcripts, reDec 25, · The transcripts obtained by RNA‐Seq were annotated with Gencode vM18 and NONCODEv5 The TPM value was used to quantify the assembled transcriptome, including differentially expressed protein‐coding and lncRNA genes (Table S4) The length of most transcripts was concentrated within 1000 nt (Figure 1A)



Hiv 1 Tat Interacts With A Kaposi S Sarcoma Associated Herpesvirus Reactivation Upregulated Antiangiogenic Long Noncoding Rna Linc And Antagonizes Its Function Journal Of Virology



Invited Review Long Non Coding Rnas Important Regulators In The Development Function And Disorders Of The Central Nervous System Cuevas Diaz Duran 19 Neuropathology And Applied Neurobiology Wiley Online Library
Using CHESS21 as the reference annotation database (containing both proteincoding and lncRNAs genes) we used a cumulative stepwise intersection method to merge it with the rest of the five lncRNAs annotation databases in this order (i) FANTOM50v3, (ii) LNCipedia52, (iii) NONCODEv50, (iv) MiTranscriptomev2 and (v) BIGTranscriptomev1 atNONCODEv5, the systematic public database that is dedicated to the collection and annotation of lncRNA, has recorded 548,640 lncRNA transcripts from 17 different species, with the expression profiles of entire human and mouse genome being annotatedJan 01, 19 · NONCODEv5, the systematic public database that is dedicated to the collection and annotation of lncRNA, has recorded 548,640 lncRNA transcripts from 17 different species, with the expression profiles of entire human and mouse genome being annotated
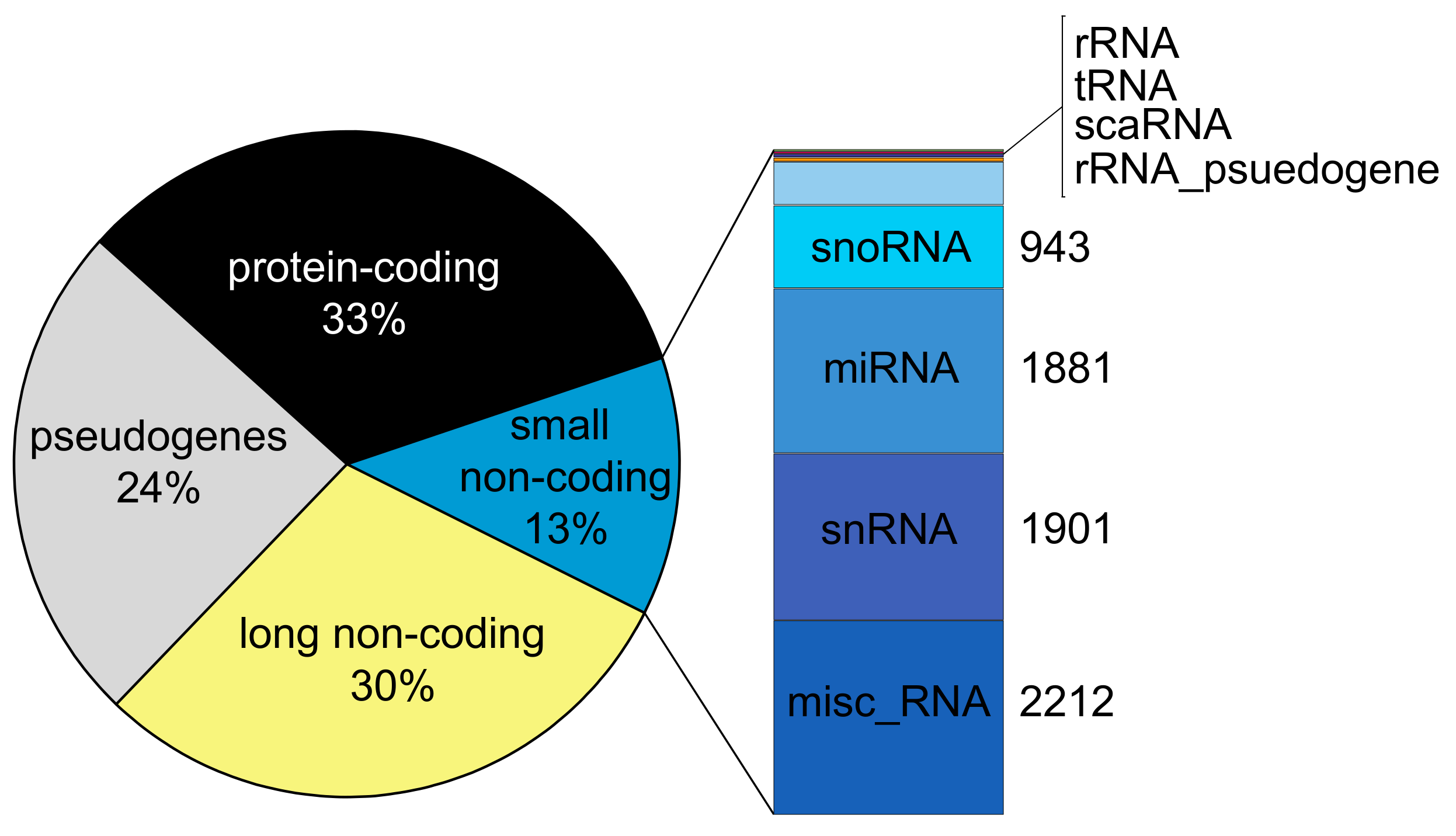


Ijms Free Full Text Non Coding Rna Signatures Of B Cell Acute Lymphoblastic Leukemia Html



Noncodev5 A Comprehensive Annotation Database For Long Non Coding Rnas Lncrna Blog
Jan 24, · The rhesus macaque is a prime model animal in neuroscience A comprehensive transcriptomic and open chromatin atlas of the rhesus macaque brain is key to a deeper understanding of the brain HereThe ncRNA database including sequence, documentation and analysisPMID NONCODEV5 a comprehensive annotation database for long noncoding RNAs PMID Structure, targetspecificity and expression of PN_LNC_N13, a long noncoding RNA differentially expressed in apomictic and sexual Paspalum notatum



Invited Review Long Non Coding Rnas Important Regulators In The Development Function And Disorders Of The Central Nervous System Cuevas Diaz Duran 19 Neuropathology And Applied Neurobiology Wiley Online Library



Lncar A Comprehensive Resource For Lncrnas From Cancer Arrays Cancer Research
Nov 06, 19 · To date, human lncRNA transcripts have been identified according to NONCODEv5 database and their number continues to increase Recent studies have demonstrated that several lncRNAs have a key regulatory role in various diseases including CRCOct 30, · Long noncoding RNAs (lncRNAs) have a size of more than 0 bp and are known to regulate a host of crucial cellular processes like proliferation, differentiation and apoptosis by regulating gene expression While small noncoding RNAs (ncRNAs) such as miRNAs, siRNAs, Piwiinteracting RNAs have been extensively studied in male germ cell development, the role of96,000 currently described, according to the NONCODEV5 database (10) Accumulating evidence has demonstrated an association between the altered expression of some lncRNAs with various cancer types These include GAS5, LINC‑PINT, MEG3, HOTAIR and MALAT1, which are involved in various lncRNA HAR1B has potential to be a predictive marker
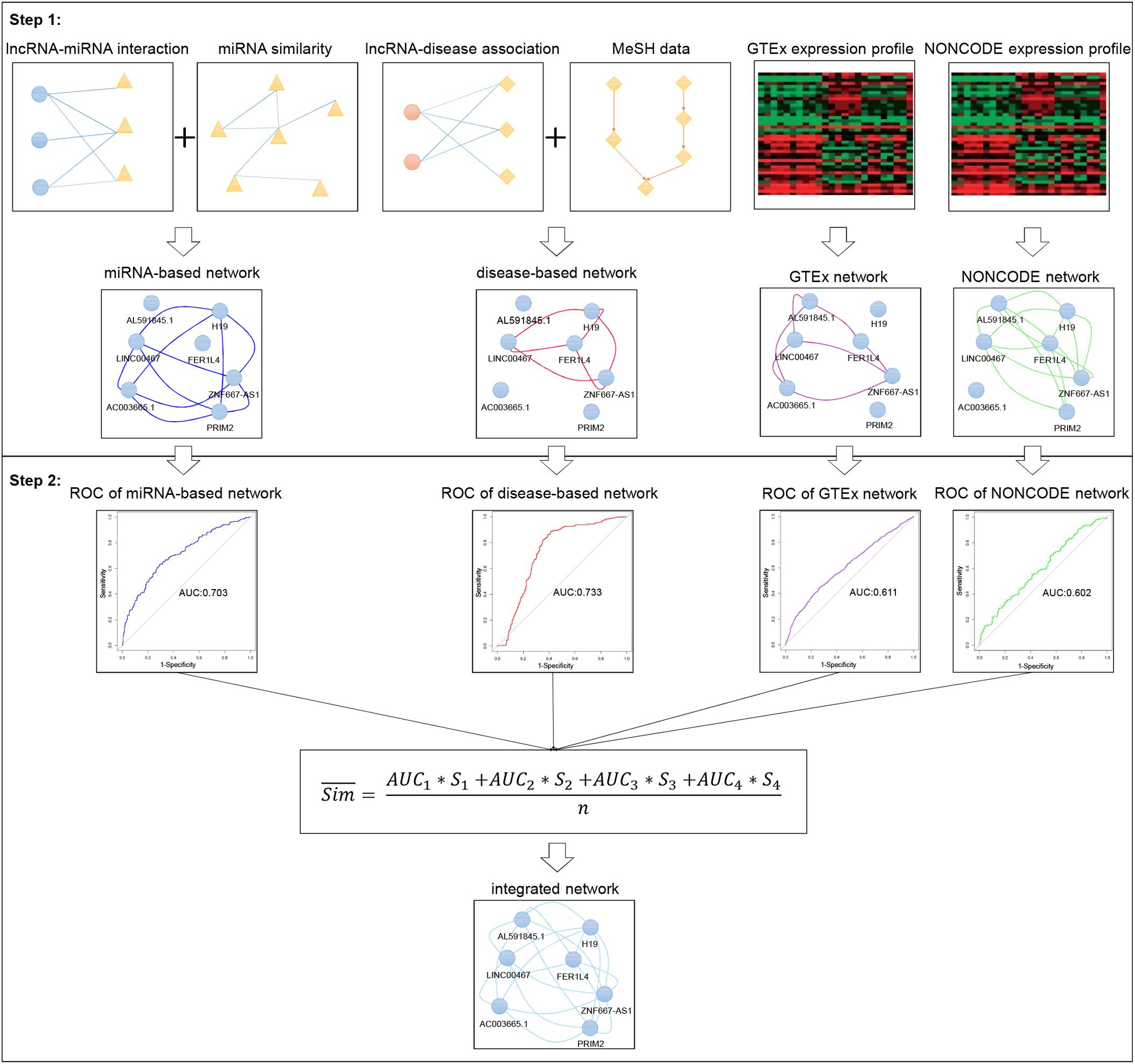


Frontiers Inferring Lncrna Functional Similarity Based On Integrating Heterogeneous Network Data Bioengineering And Biotechnology



Noncodev5 A Comprehensive Annotation Database For Long Non Coding Rnas Lncrna Blog
3 Volders PJ, Verheggen K, Menschaert G, Vandepoele K, Martens L, Vandesompele J, Mestdagh P An update on LNCipedia a database for annotated human lncRNA sequencesAccording to the NONCODE (noncodeorg) database file "NONCODEv5_human_hg38_lncRNAgtf", 172,216 lncRNA transcripts can theoretically be found in humans From our literature screening in January , we found hundreds of them potentially associated with COPD, asthma, IPF, orFeb 01, 18 · The number of lncRNAs in NONCODEv5 increased from 527 336 to 548 640
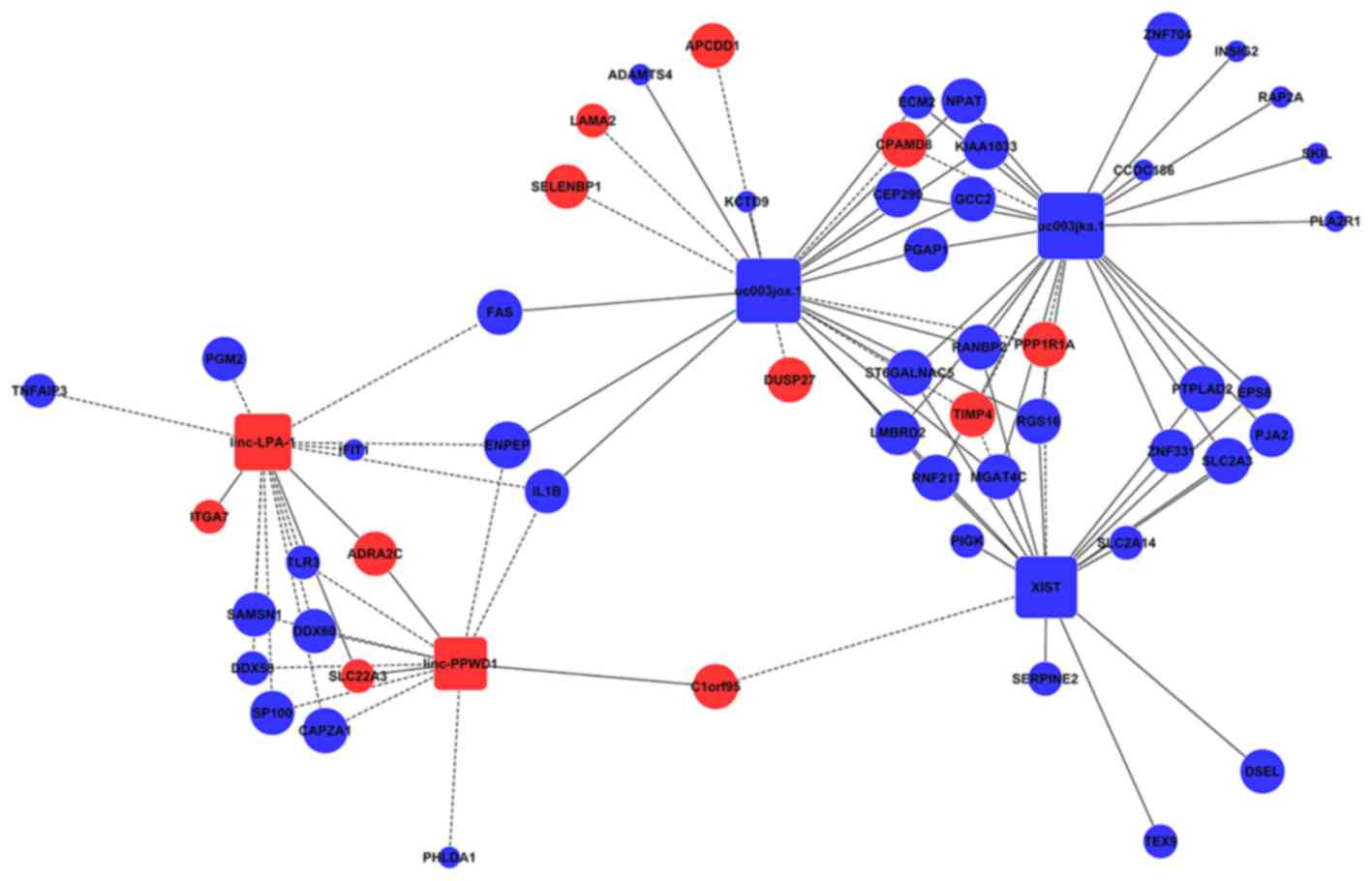


Microarray Analysis Of Long Non Coding Rna Expression Profiles In Marfan Syndrome



Lncrnakb A Comprehensive Knowledgebase Of Long Non Coding Rnas Biorxiv
Jul 05, 19 · Loop is the open research network that increases the discoverability and impact of researchers and their work Loop enables you to stay uptodate with the latest discoveries and news, connect with researchers and form new collaborationsProteincoding transcript sequences CHR Nucleotide sequences of coding transcripts on the reference chromosomesTo satisfying researchers quality demand, NONCODEv5 provides a subset searching interface You can choose the following conditions to get the high quality subset source of the data, for instance, from literature, or from RefSeq or Ensembl or GENCODE or lncRNAdb, or from the thirdgeneration sequencing technology, etc exon number of a transcript



Roles Of Lncrnas In Rice Advances And Challenges Sciencedirect



Lncar A Comprehensive Resource For Lncrnas From Cancer Arrays Cancer Research
Of these, >15,000 lncRNA genes have been annotated by the GENCODE Consortium (version 27) 13 Such identification and characterization of lncRNAs mostly rely on advanced highthroughput nextgeneration sequencing (NGS) techNONCODE NONCODE (current version v50) is an integrated knowledge database dedicated to noncoding RNAs (excluding tRNAs and rRNAs) Now, there are 17 species in NONCODE (human, mouse, cow, rat, chicken, fruitfly, zebrafish, celegans, yeast)Content Regions Description Download;
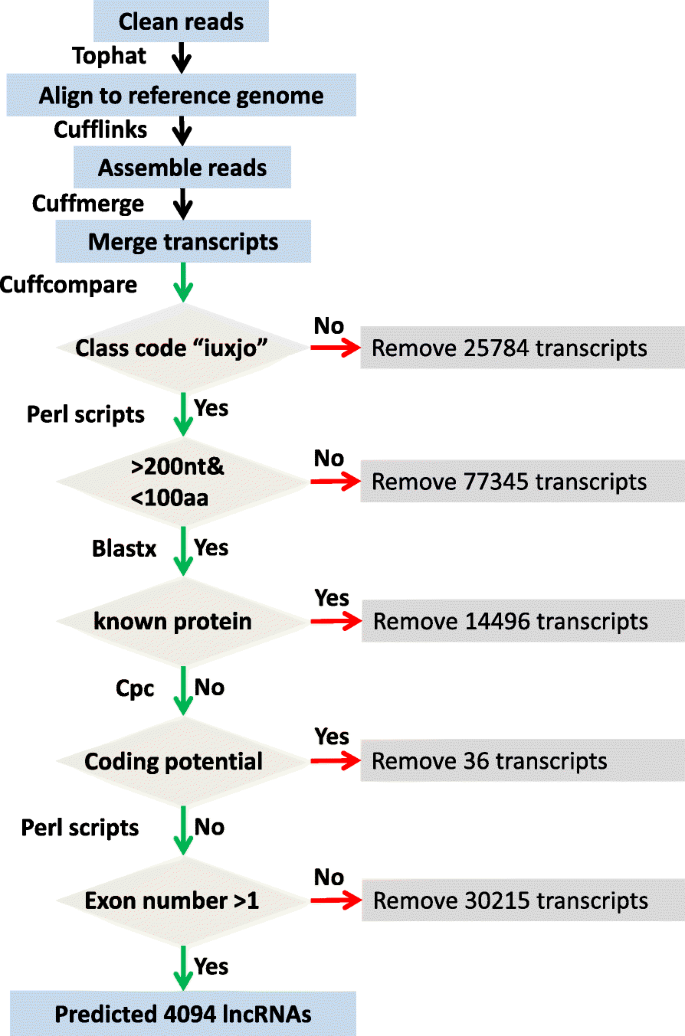


Identification And Analysis Of Long Non Coding Rnas In Response To H5n1 Influenza Viruses In Duck Anas Platyrhynchos Bmc Genomics Full Text


Noncode
Background Long noncoding RNAs (lncRNAs) are important component of mammalian genomes, where their numbers are even larger than that of proteincoding genes For example, human (Homo sapiens) (96,308 vs ,376) and mouse (Mus musculus) (87,774 vs 22,630) have more lncRNA genes than proteincoding genes in the NONCODEv5 databaseNONCODEV5 a comprehensive annotation database for long noncoding RNAs Nucleic Acids Res 18;LncRNAs, identified by NONCODEv5, were defined at a detectable cutoff of 6 reads in at least 50% of the samples in either group, resulting in 15 695 lncRNAs Fold‐change and false discovery rate of the differentially expressed lncRNAs were calculated using the R/Bioconductor packages edgeR and limma/variance modeling at the observational level



The How And Why Of Lncrna Function An Innate Immune Perspective Sciencedirect
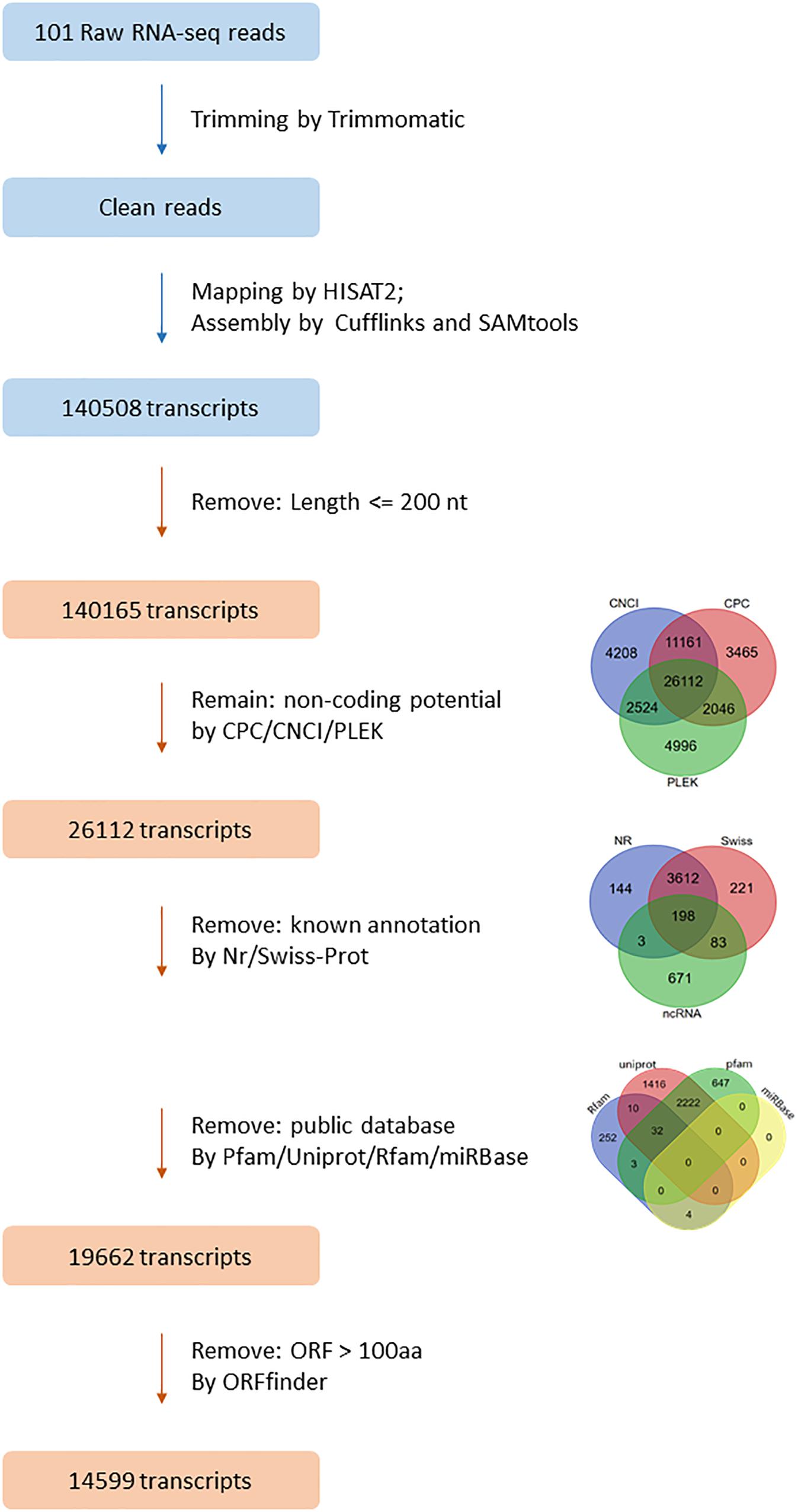


Frontiers Identification And Expression Analysis Of Long Non Coding Rna In Large Yellow Croaker Larimichthys Crocea In Response To Cryptocaryon Irritans Infection Genetics
46D308–D314 doi /nar/gkx1107 Crossref Medline Google Scholar;
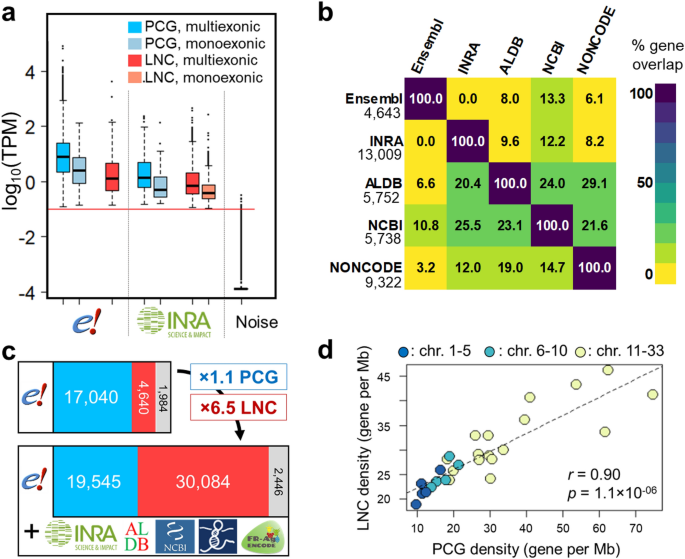


An Integrative Atlas Of Chicken Long Non Coding Genes And Their Annotations Across 25 Tissues Scientific Reports


Noncode



Computational Methods And Applications For Identifying Disease Associated Lncrnas As Potential Biomarkers And Therapeutic Targets Molecular Therapy Nucleic Acids
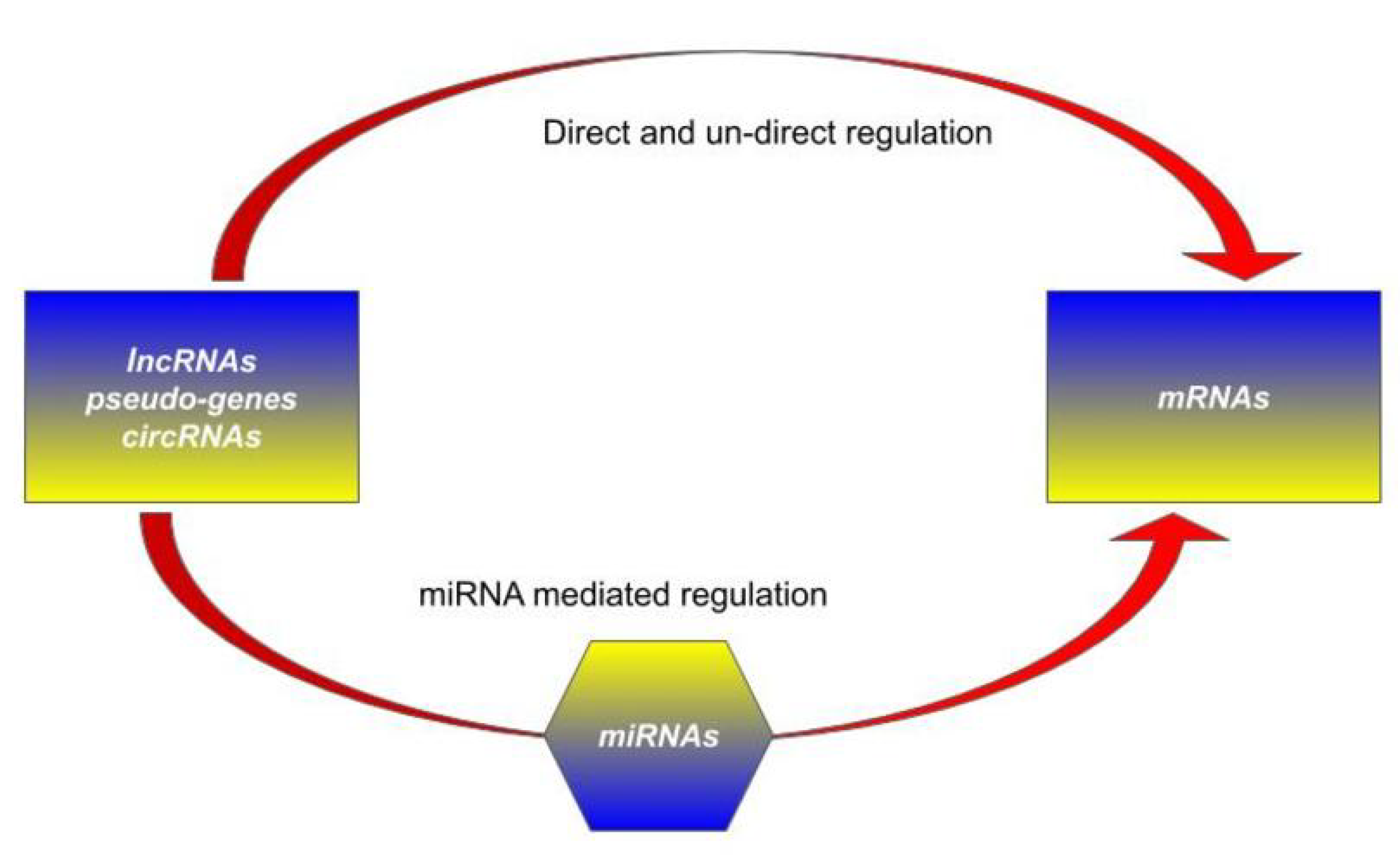


Cells Free Full Text Competing Endogenous Rnas Non Coding Rnas And Diseases An Intertwined Story Html


A Noninvasive Prediction Nomogram For Lymph Node Metastasis Of Hepatocellular Carcinoma Based On Serum Long Noncoding Rnas Document Gale Academic Onefile



Computational Methods And Applications For Identifying Disease Associated Lncrnas As Potential Biomarkers And Therapeutic Targets Molecular Therapy Nucleic Acids


Noncode



Genes Free Full Text Prelnc An Accurate Tool For Predicting Lncrnas Based On Multiple Features Html


Lncrnakb Methods
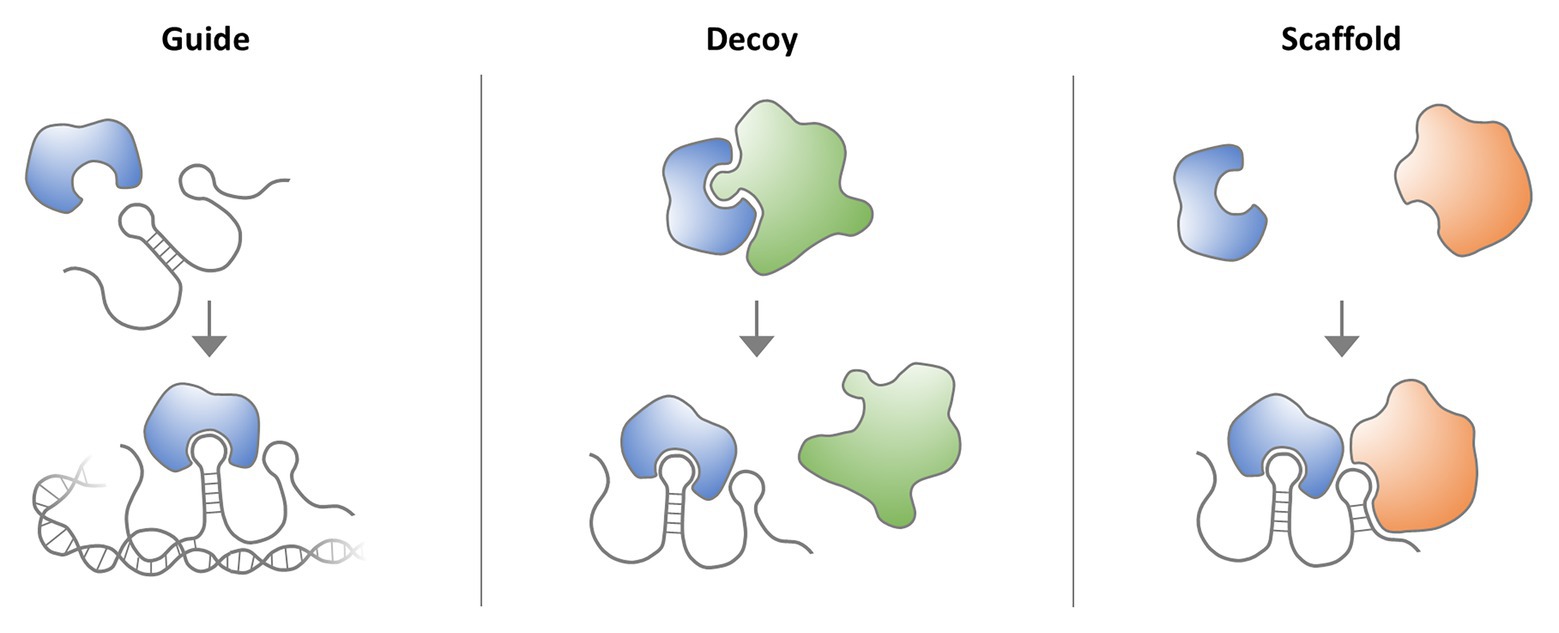


Frontiers Disease Causing Mutations And Rearrangements In Long Non Coding Rna Gene Loci Genetics


Noncode



Noncodev5 A Comprehensive Annotation Database For Long Non Coding Rnas Lncrna Blog



Functional Analysis Of Transcription Factors And Genes In The Download Scientific Diagram



Noncode Database Commons


Lncrnakb Methods


Noncode


Academic Oup Com Nar Article Pdf 49 D1 D165 Gkaa1046 Pdf



Lncrna Comparison With The Noncodev5 Database Based On Sequence Similarity Download Table



Lncrnakb A Comprehensive Knowledgebase Of Long Non Coding Rnas Biorxiv


Annolnc2 The One Stop Portal To Systematically Annotate Novel Lncrnas For Human And Mouse Gao Lab


Lncrnakb Methods
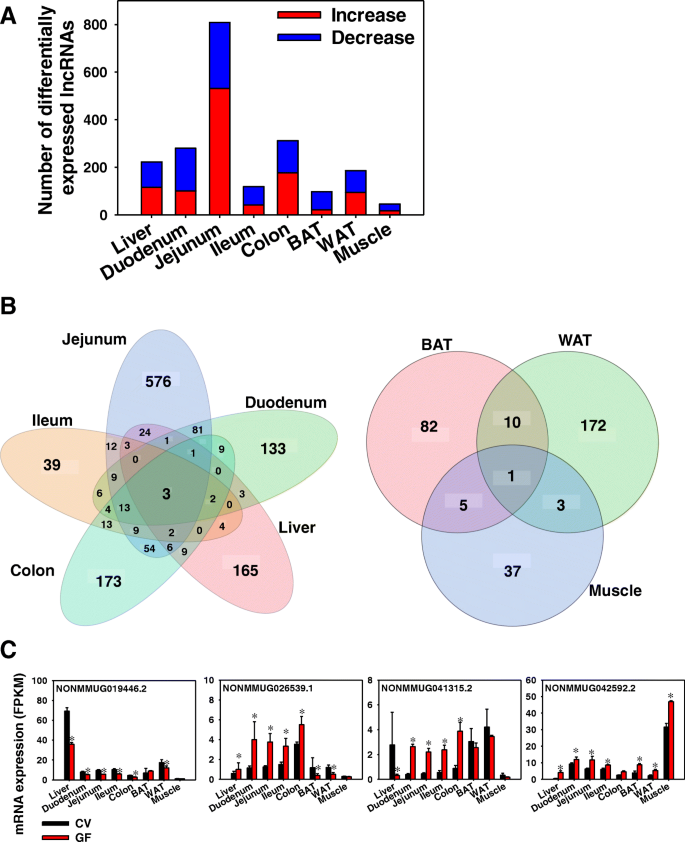


Coordinate Regulation Of Long Non Coding Rnas And Protein Coding Genes In Germ Free Mice Bmc Genomics Full Text
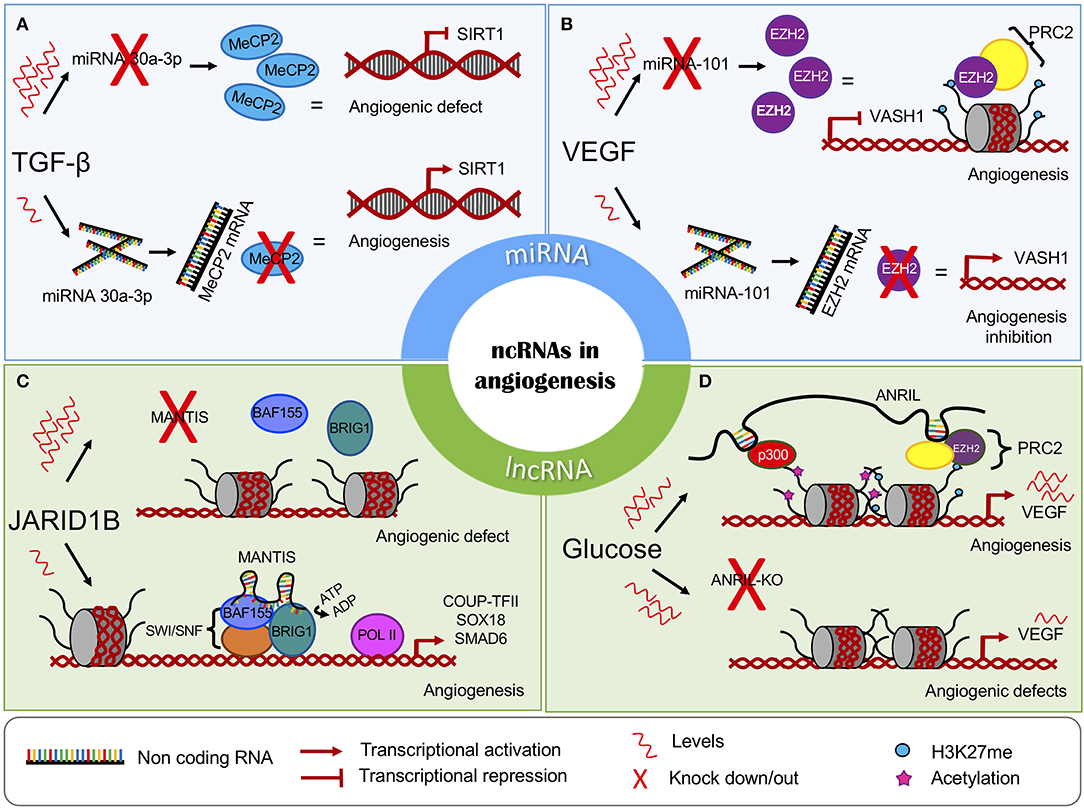


Frontiers The Regulatory Roles Of Non Coding Rnas In Angiogenesis And Neovascularization From An Epigenetic Perspective Oncology


Lncrna Expression Profile During Autophagy And Malat1 Function In Macrophages


Noncode



Database Lncrna Blog


Gale Academic Onefile Document Prelnc An Accurate Tool For Predicting Lncrnas Based On Multiple Features


Noncodev5强势来袭 一款全面的ncrnas注释数据库 企业动态 洛阳晶云信息科技有限公司



Noncodev5 A Comprehensive Annotation Database For Long Non Coding Rnas Lncrna Blog
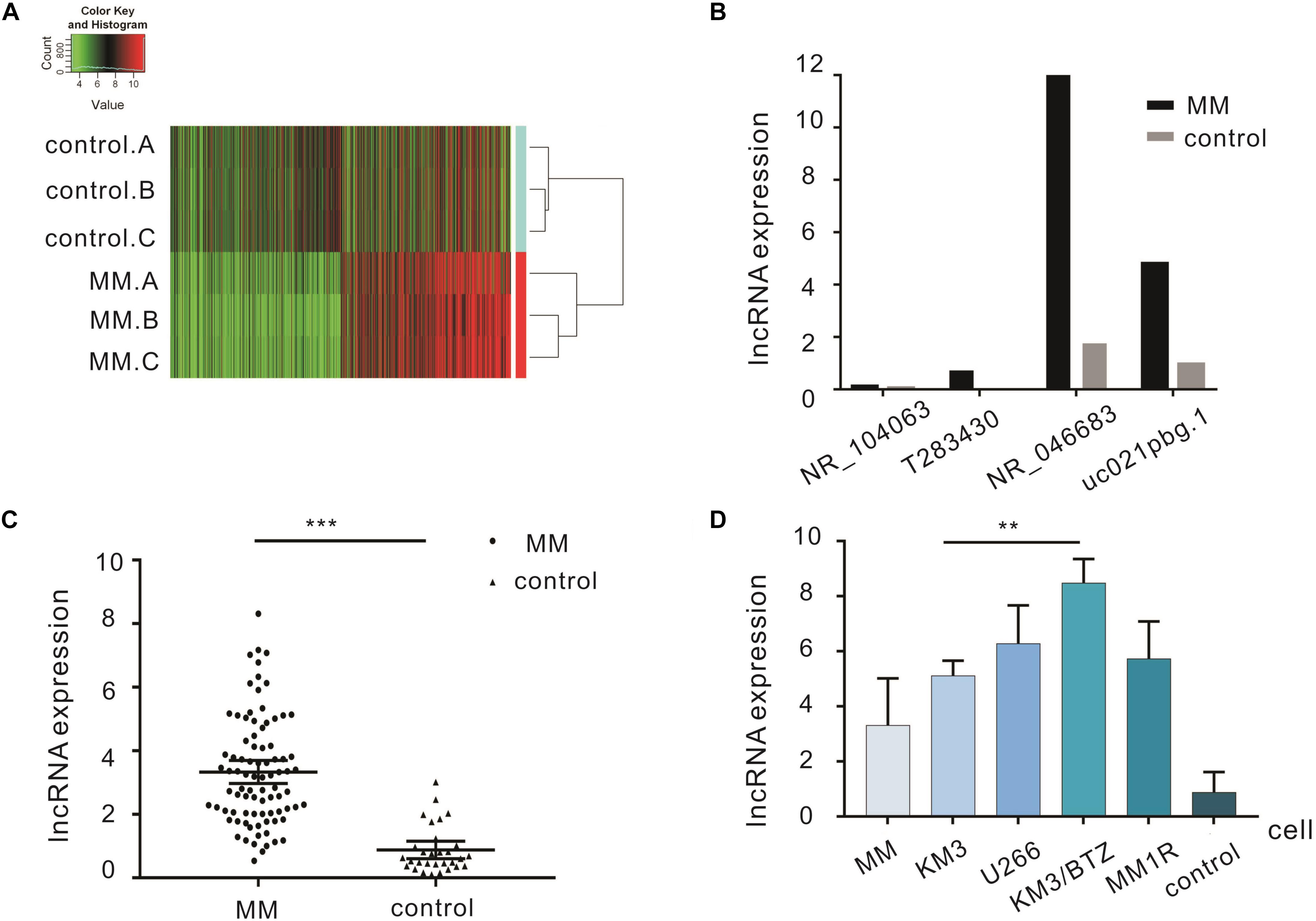


Frontiers Upregulation Of Lncrna Nr 0466 Serves As A Prognostic Biomarker And Potential Drug Target For Multiple Myeloma Pharmacology


Noncode


Full Article Lncrnas In Adaptive Immunity Role In Physiological And Pathological Conditions
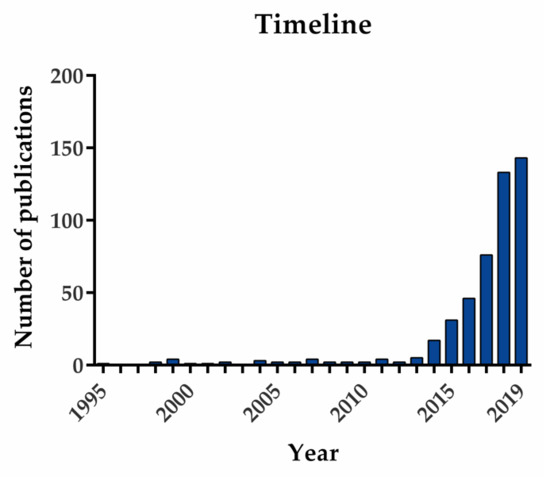


Cancers Free Full Text The Challenges And Opportunities Of Lncrnas In Ovarian Cancer Research And Clinical Use Html
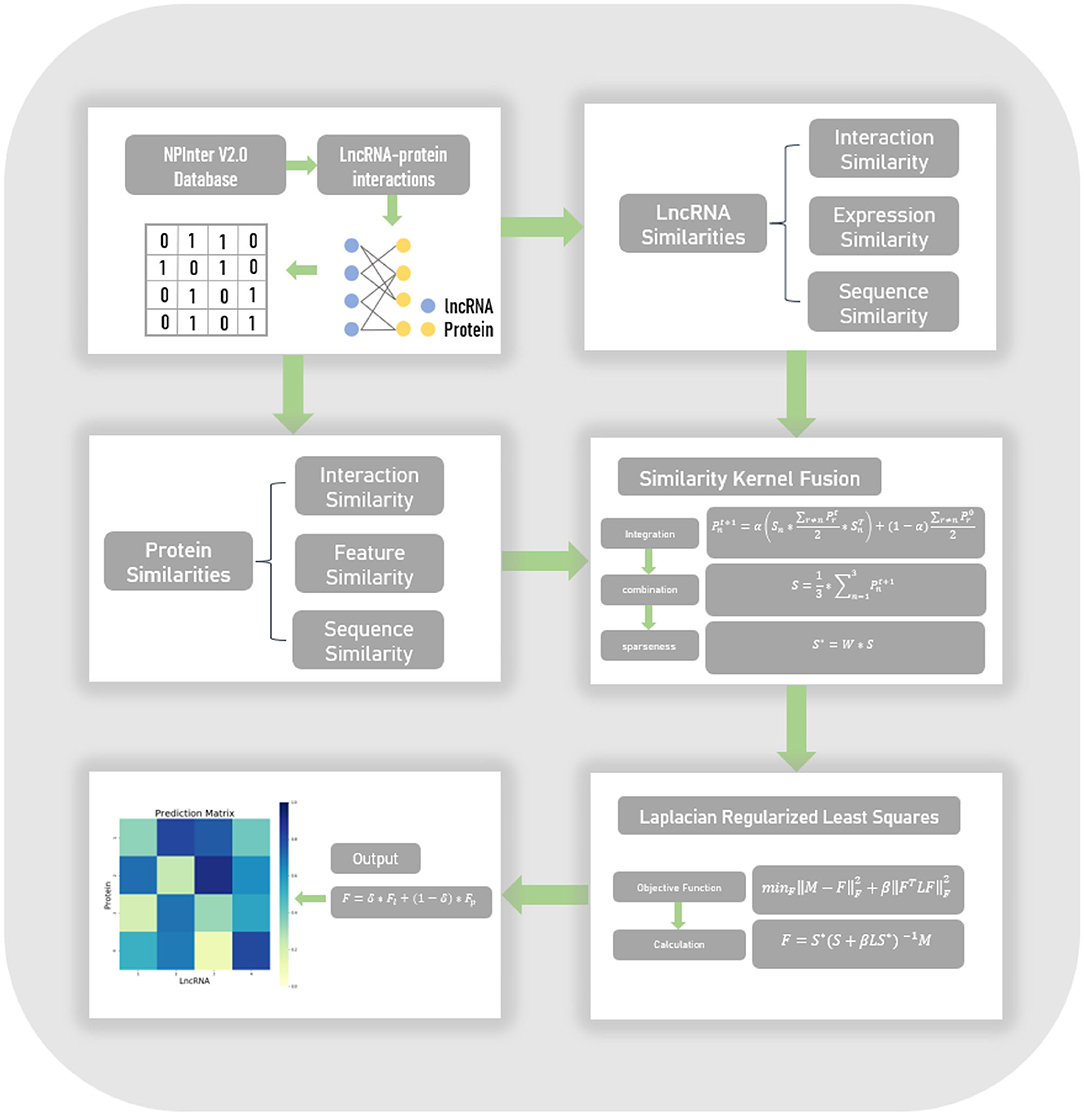


Frontiers Lpi Skf Predicting Lncrna Protein Interactions Using Similarity Kernel Fusions Genetics
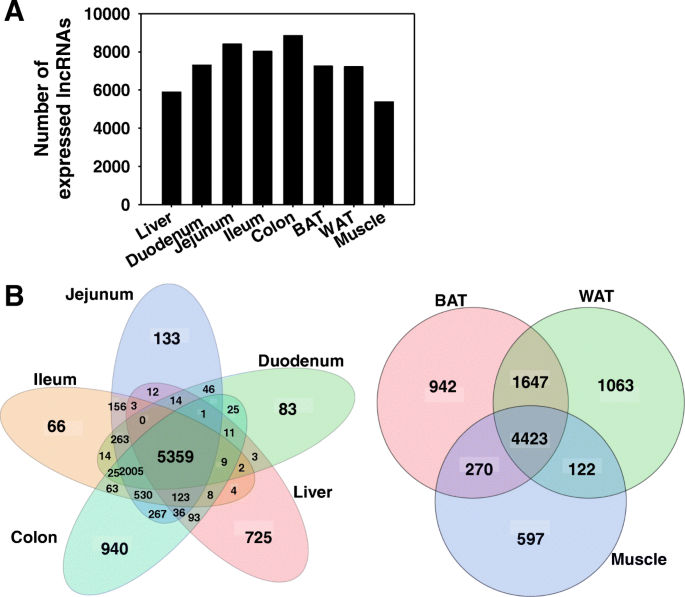


Coordinate Regulation Of Long Non Coding Rnas And Protein Coding Genes In Germ Free Mice Bmc Genomics Full Text
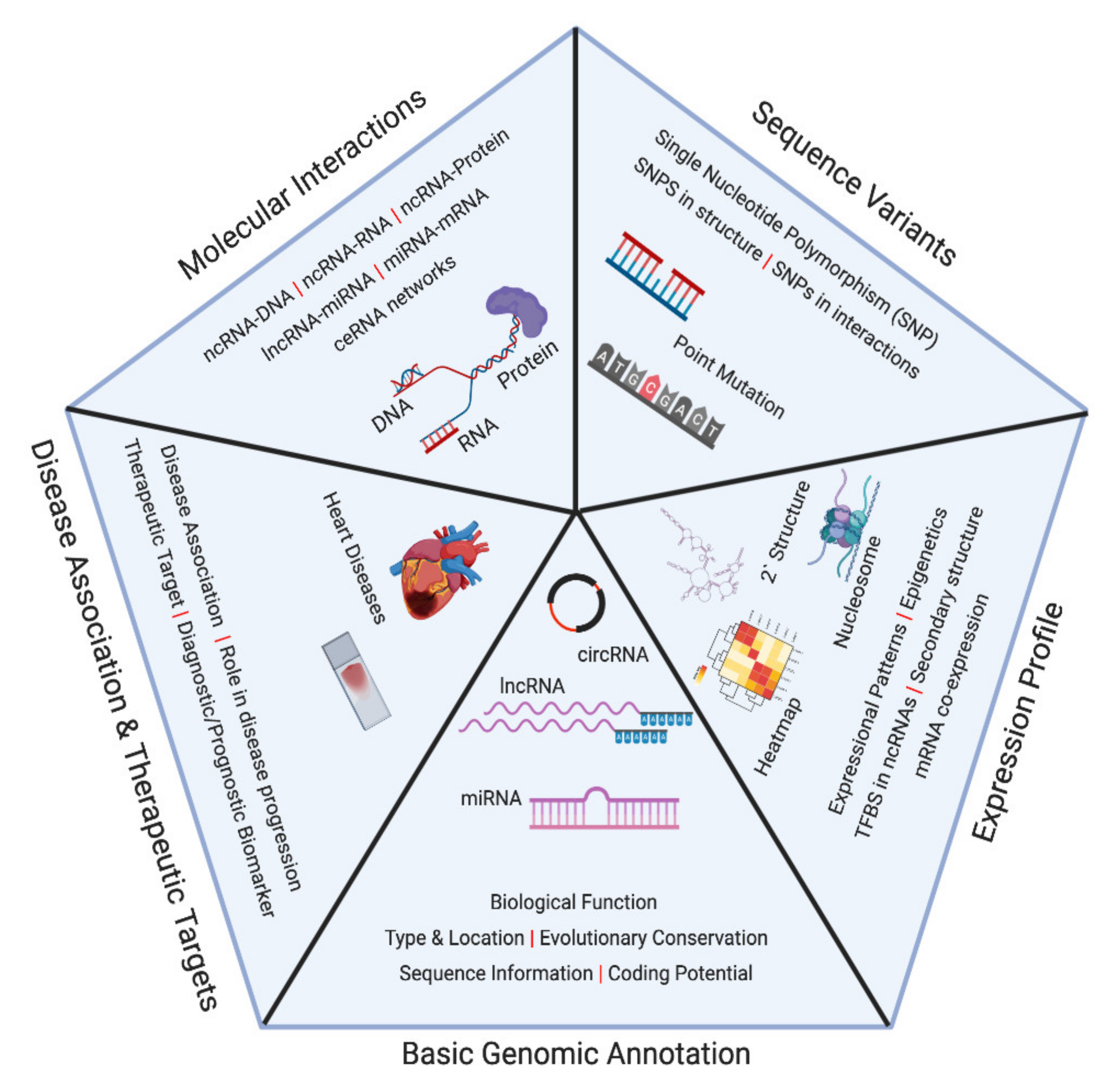


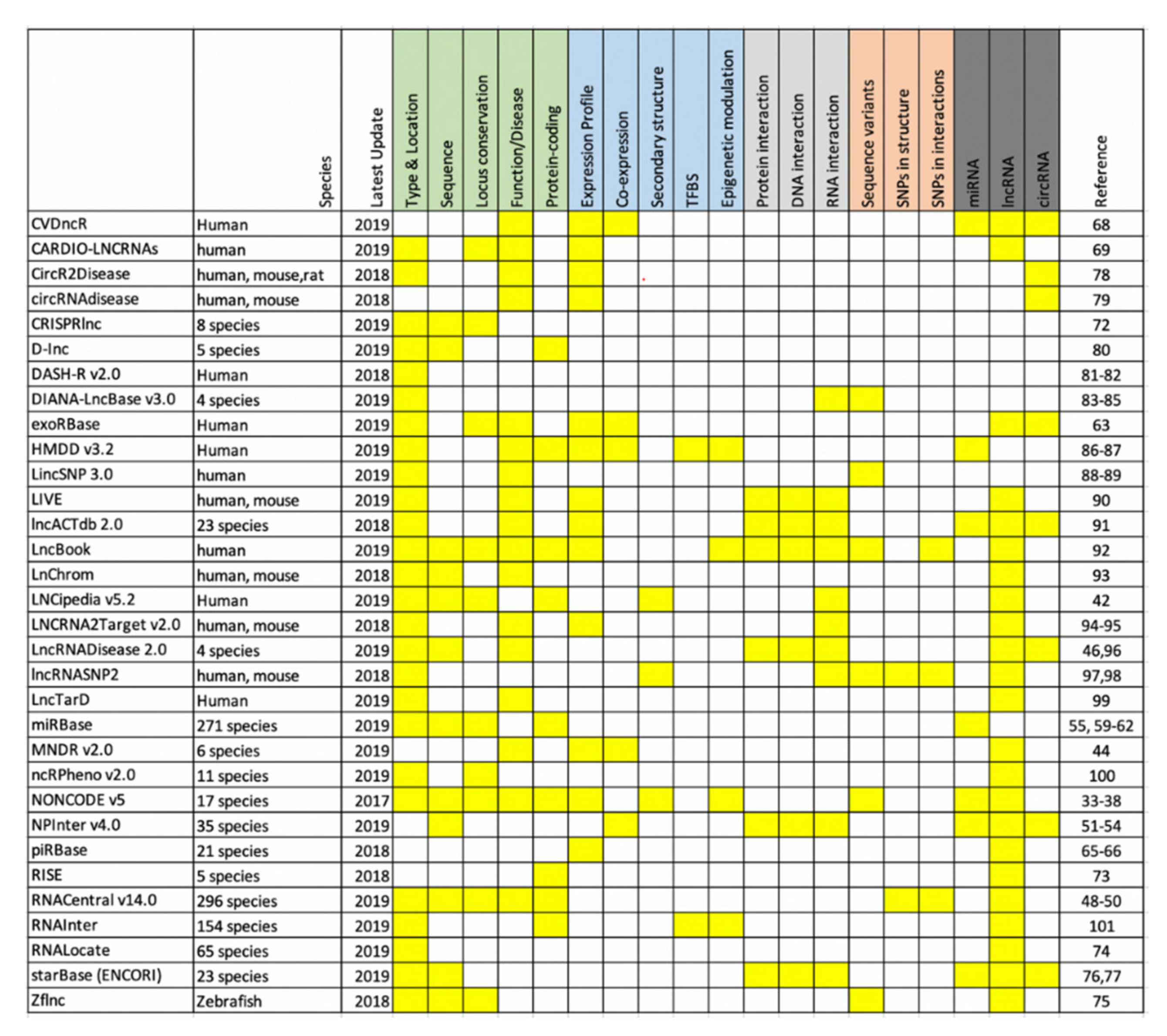
Ncrna Free Full Text Non Coding Rna Databases In Cardiovascular Research Html
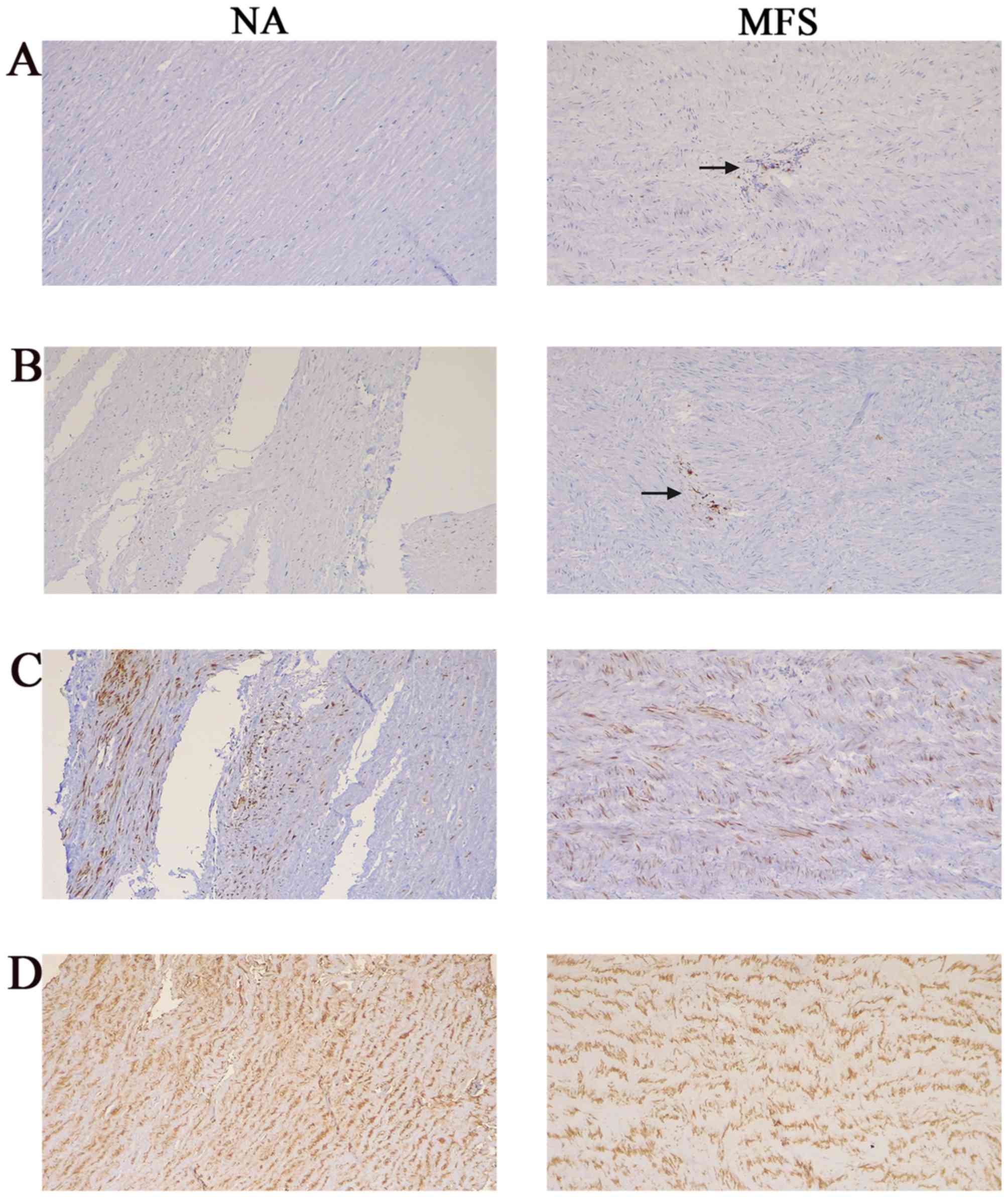


Microarray Analysis Of Long Non Coding Rna Expression Profiles In Marfan Syndrome


Pdfs Semanticscholar Org F791 Addf2a5cf7adf3b52b3ba26f0 Pdf



One Locus With Two Roles Microrna Independent Functions Of Microrna Host Gene Locus Encoded Long Noncoding Rnas Sun Wires Rna Wiley Online Library



Pdf Prelnc An Accurate Tool For Predicting Lncrnas Based On Multiple Features Semantic Scholar



Meg8 Long Noncoding Rna Contributes To Epigenetic Progression Of The Epithelial Mesenchymal Transition Of Lung And Pancreatic Cancer Cells Journal Of Biological Chemistry



Lncrna Comparison With The Noncodev5 Database Based On Sequence Similarity Download Table



Pdf Noncodev5 A Comprehensive Annotation Database For Long Non Coding Rnas



Computational Methods And Applications For Identifying Disease Associated Lncrnas As Potential Biomarkers And Therapeutic Targets Molecular Therapy Nucleic Acids
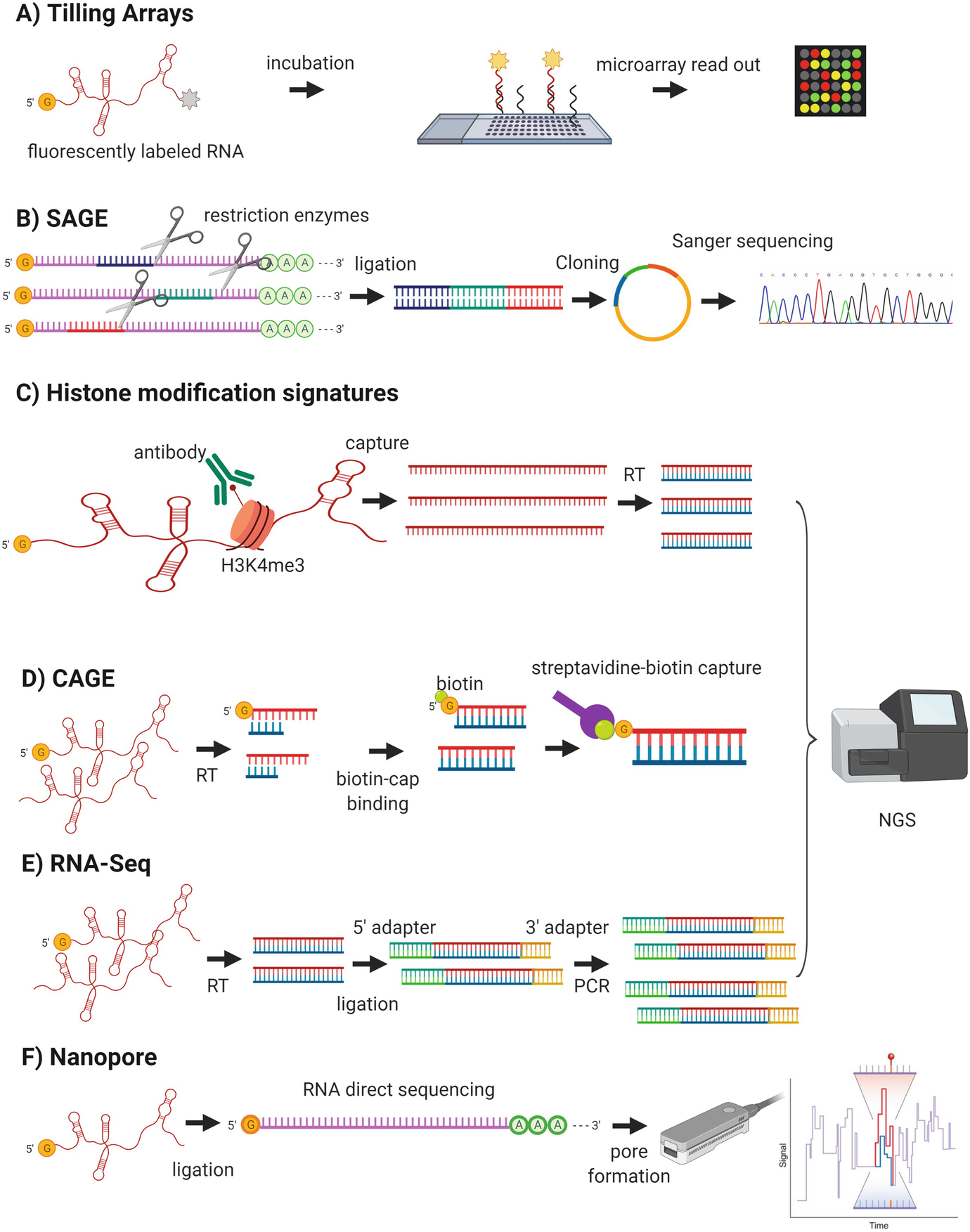


Long Non Coding Rnas Diversity In Form And Function From Microbes To Humans Springerlink



Lncrna Comparison With The Noncodev5 Database Based On Sequence Similarity Download Table



The Computational Pipeline Used To Identify Lncrna Genes From Rna Seq Download Scientific Diagram
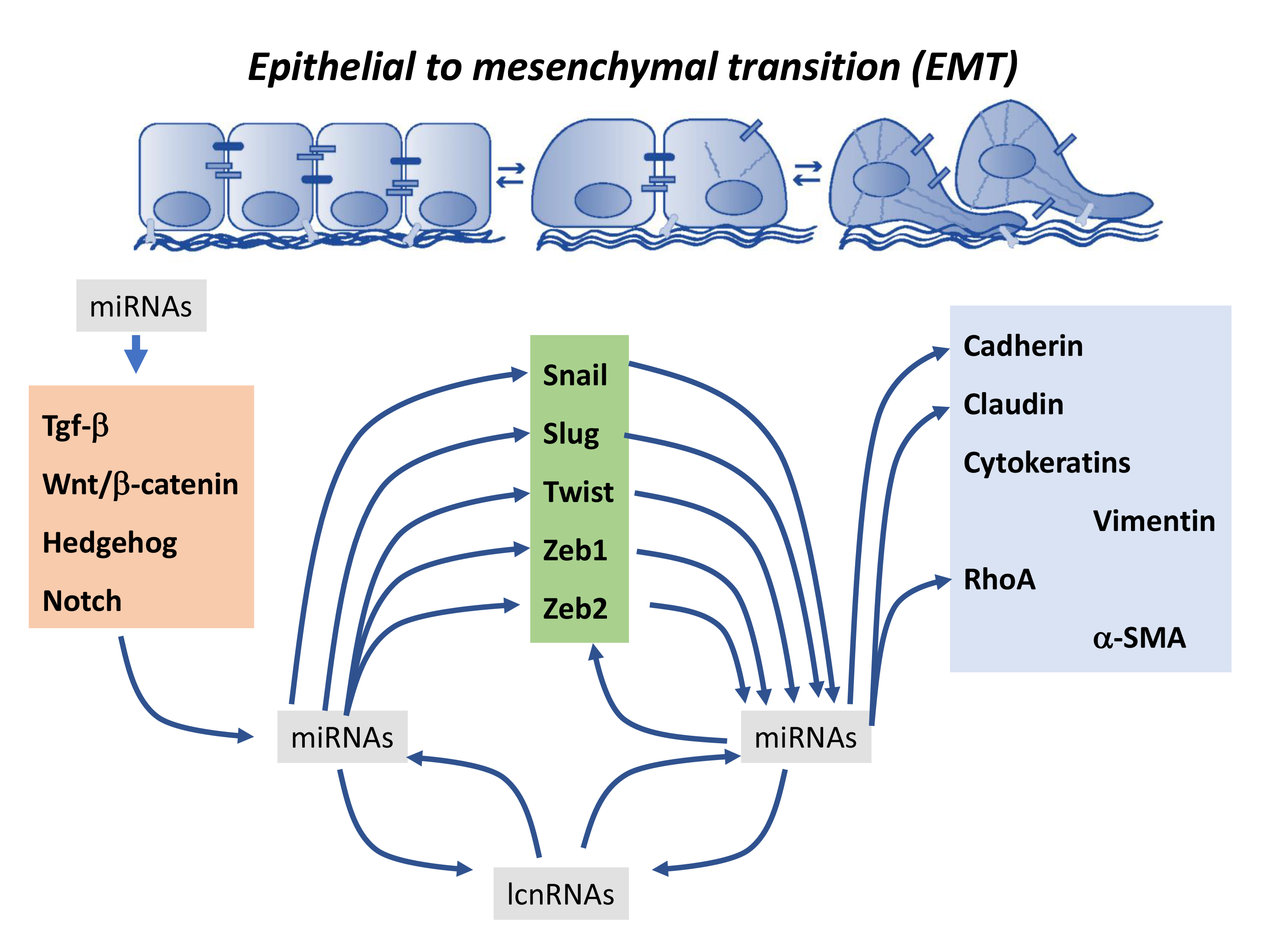


Ncrna Free Full Text Functional Role Of Non Coding Rnas During Epithelial To Mesenchymal Transition Html
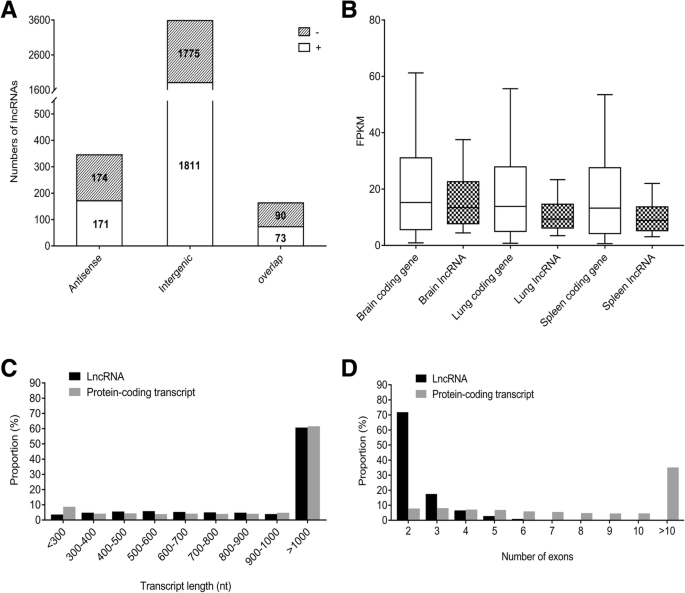


Identification And Analysis Of Long Non Coding Rnas In Response To H5n1 Influenza Viruses In Duck Anas Platyrhynchos Bmc Genomics Full Text



Noncodev5 A Comprehensive Annotation Database For Long Non Coding Rnas Lncrna Blog
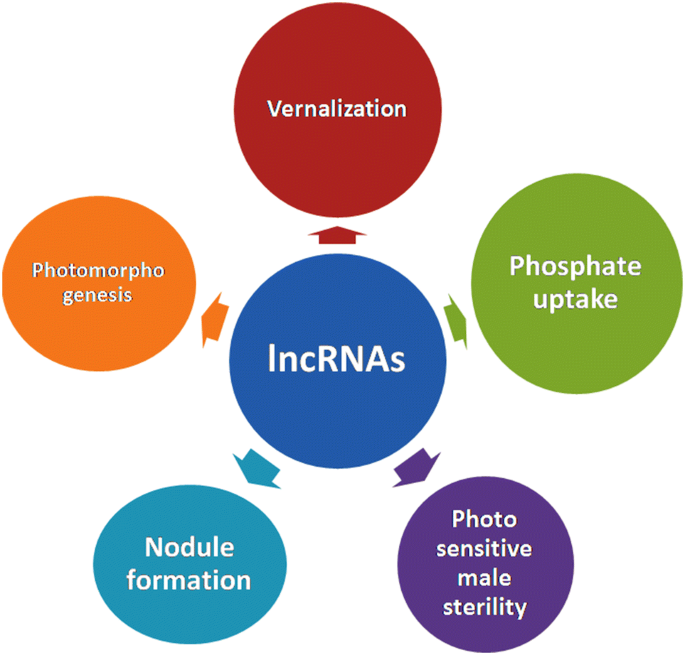


Updates On Plant Long Non Coding Rnas Lncrnas The Regulatory Components Springerlink
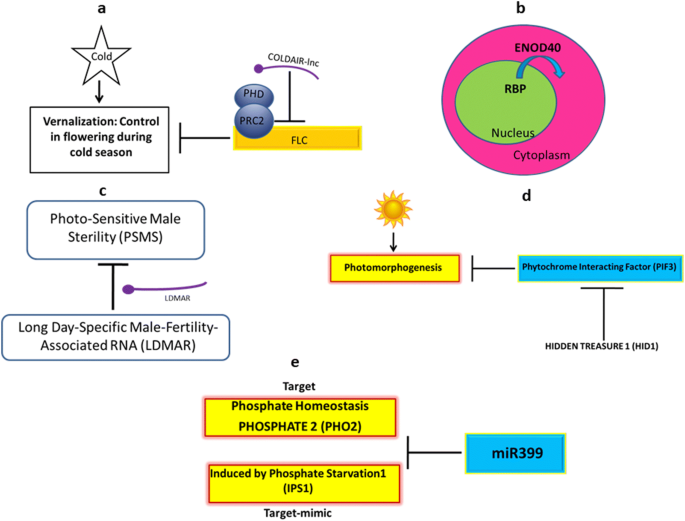


Updates On Plant Long Non Coding Rnas Lncrnas The Regulatory Components Springerlink
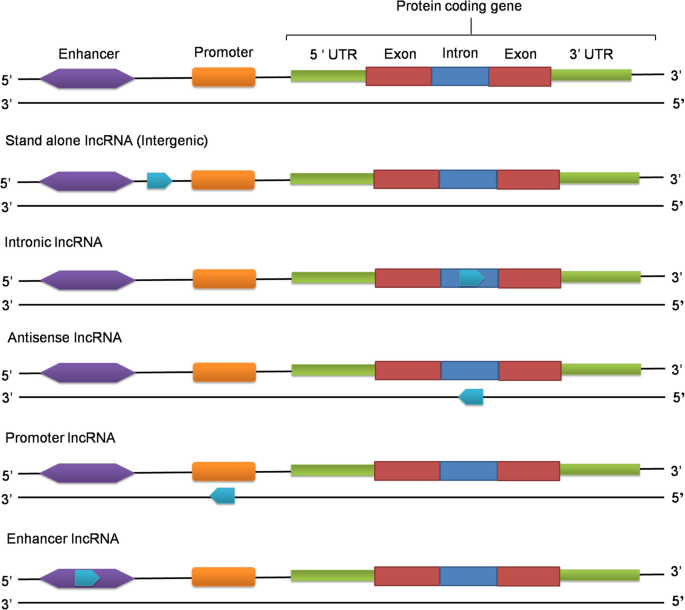


Long Non Coding Rnas Lncrnas In Spermatogenesis And Male Infertility Reproductive Biology And Endocrinology Full Text



Roles Of Lncrnas In Rice Advances And Challenges Sciencedirect


Lncrnakb Methods
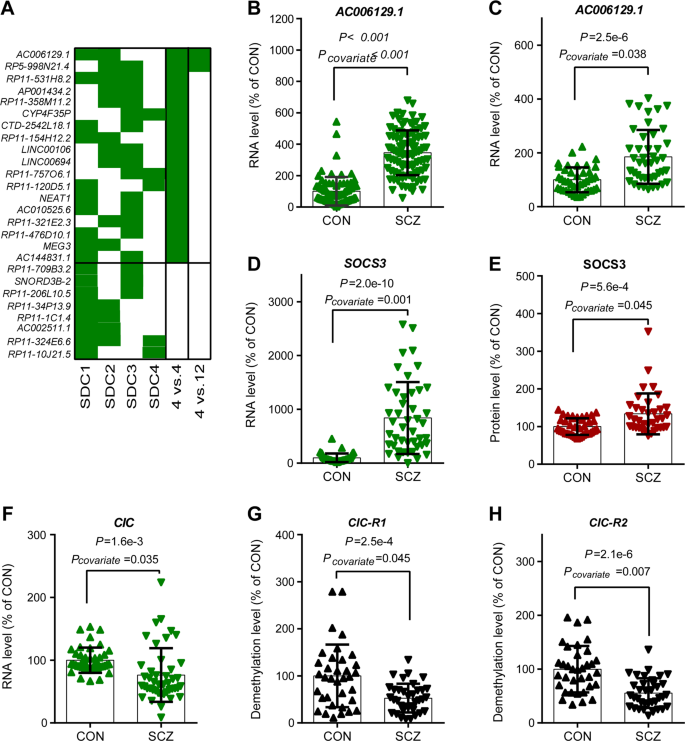


Lncrna Ac 1 Reactivates A Socs3 Mediated Anti Inflammatory Response Through Dna Methylation Mediated Cic Downregulation In Schizophrenia Molecular Psychiatry



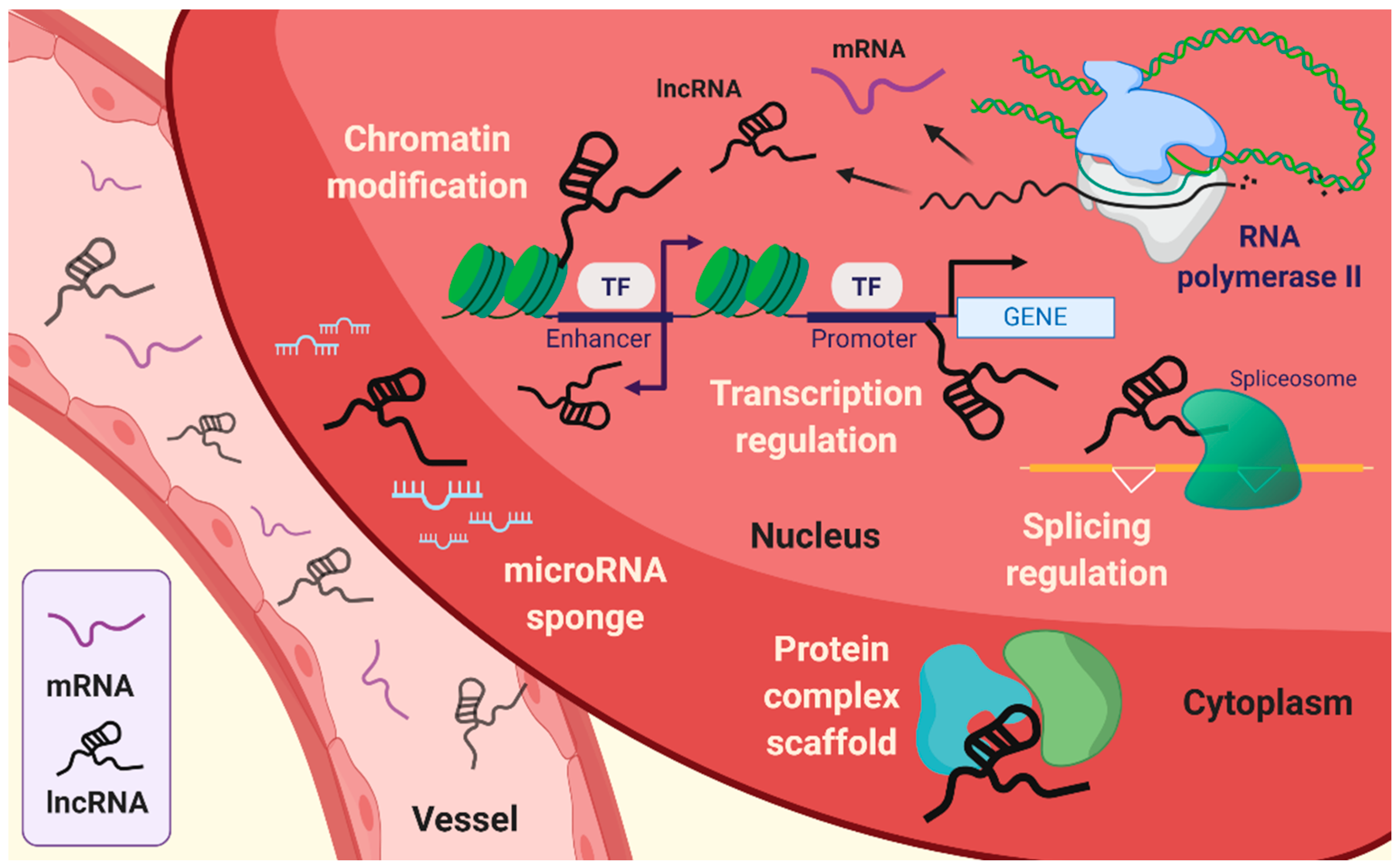
Lncrnas Coming Of Age Circulation Research



Single Cell Long Non Coding Rna Landscape Of T Cells In Human Cancer Immunity Biorxiv



Pdf Noncodev5 A Comprehensive Annotation Database For Long Non Coding Rnas



Lncrnakb A Comprehensive Knowledgebase Of Long Non Coding Rnas Biorxiv


2



Identification Functional Prediction And Key Lncrna Verification Of Cold Stress Related Lncrnas In Rats Liver Scientific Reports



Lncrna Hrceg Regulated By Hdac1 Inhibits Cells Proliferation And Epithelial Mesenchymal Transition In Gastric Cancer Cancer Genetics



Lncrna Comparison With The Noncodev5 Database Based On Sequence Similarity Download Table


Noncodev5 A Comprehensive Annotation Database For Long Non Coding Rnas Lncrna Blog


Noncodev5强势来袭 一款全面的ncrnas注释数据库 企业动态 洛阳晶云信息科技有限公司



The Role Of Long Non Coding Rnas In Cardiac Development And Disease Abstract Europe Pmc



Long Non Coding Rna Review And Implications In Acute Lung Inflammation Sciencedirect


Academic Oup Com Nar Article Pdf 49 D1 D165 Gkaa1046 Pdf


Lncrna Expression Profile During Autophagy And Malat1 Function In Macrophages
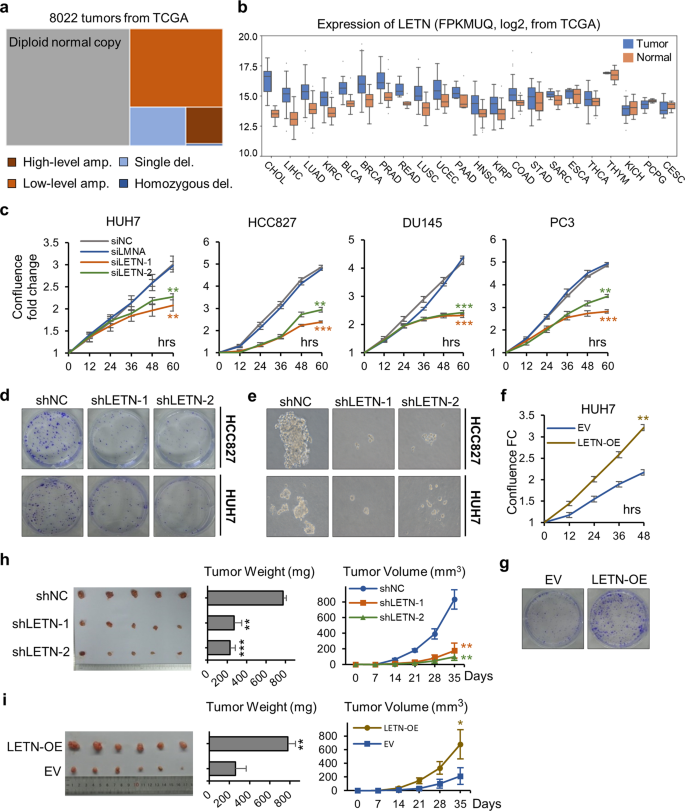


Mutual Dependency Between Lncrna Letn And Protein Npm1 In Controlling The Nucleolar Structure And Functions Sustaining Cell Proliferation Cell Research



Functional Long Non Coding Rnas In Hepatocellular Carcinoma Sciencedirect



Lncrna Expression Predicts Mrna Abundance Epigenomics


Integrated Analysis Of Long Non Coding Rnas And Mrna Profiles Reveals Potential Sex Dependent Biomarkers Of Bevacizumab Erlotinib Response In Advanced Lung Cancer


